Connectomics
!pip install --upgrade vfb_connect
!pip install --upgrade git+git://github.com/schlegelp/navis@master
!pip install --upgrade git+git://github.com/schlegelp/pymaid@master
!pip install --upgrade ipywidgets
!pip install --upgrade jsonpath_rw
!pip install --upgrade seaborn
!pip install --upgrade neuprint-python
!pip install --upgrade flybrains
# Import libs and initialise API objects
from vfb_connect.cross_server_tools import VfbConnect
import pandas as pd
vc = VfbConnect()
import pymaid
import navis
navis.set_pbars(jupyter=False)
pymaid.set_pbars(jupyter=False)
# Connect to the VFB CATMAID server hosting the FAFB data
rm = pymaid.connect_catmaid(server="https://fafb.catmaid.virtualflybrain.org/", api_token=None, max_threads=10)
# Test call to see if connection works
print(f'Server is running CATMAID version {rm.catmaid_version}')
WARNING: Could not load OpenGL library.
INFO : Global CATMAID instance set. Caching is ON. (pymaid)
Server is running CATMAID version 2020.02.15-905-g93a969b37
VFB provides a simple methods for exploring connectomics data
get_connected_neurons_by_type allows queries for connections between defined upstream and downstream neuron types/classes. This supports a level of abstraction not available without VFB, where grouping of neurons by higher level classifications is limited to extraction of classification from standardised names. The following examples take advantage of the abstract clasifications on VFB to explore direct synaptic connections.
# Find all GABA-regic inputs onto descending neurons
vc.get_connected_neurons_by_type(upstream_type='GABAergic neuron',
downstream_type='adult descending neuron',
weight=10).sort_values('weight', ascending=False, ignore_index=True)
# Get connections between pacemaker neurons
vc.get_connected_neurons_by_type(upstream_type='pacemaker neuron',
downstream_type='pacemaker neuron',
weight=10).sort_values('weight', ascending=False)
| upstream_neuron_id | upstream_neuron_name | weight | downstream_neuron_id | downstream_neuron_name | upstream_class | downstream_class | up_data_source | up_accession | down_source | down_accession | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 28 | VFB_jrchjtex | DN1a_R - 264083994 | 79 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1a neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 264083994 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 26 | VFB_jrchjtey | DN1a_R - 5813022274 | 75 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1a neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 5813022274 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 23 | VFB_jrchjtey | DN1a_R - 5813022274 | 63 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1a neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 5813022274 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 22 | VFB_jrchjtex | DN1a_R - 264083994 | 55 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1a neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 264083994 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 3 | VFB_jrchk089 | LPN_R - 480029788 | 43 | VFB_jrchk08a | LPN_R - 450034902 | LP neuron | LP neuron | neuprint_JRC_Hemibrain_1point1 | 480029788 | neuprint_JRC_Hemibrain_1point1 | 450034902 |
| 7 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | 40 | VFB_jrchjzxy | LNd_R - 5813069648 | s-LNv neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 511051477 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 1 | VFB_jrchjzxy | LNd_R - 5813069648 | 37 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | LNd neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 5 | VFB_jrchjtf0 | DN1pA_R - 324846570 | 37 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 324846570 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 16 | VFB_jrchjtf2 | DN1pA_R - 387944118 | 34 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 387944118 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 20 | VFB_jrchjtf1 | DN1pA_R - 325529237 | 33 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1p neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 325529237 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 29 | VFB_jrchjtf3 | DN1pA_R - 387166379 | 30 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 387166379 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 11 | VFB_jrchjtf1 | DN1pA_R - 325529237 | 30 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 325529237 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 21 | VFB_jrchk08a | LPN_R - 450034902 | 30 | VFB_jrchk089 | LPN_R - 480029788 | LP neuron | LP neuron | neuprint_JRC_Hemibrain_1point1 | 450034902 | neuprint_JRC_Hemibrain_1point1 | 480029788 |
| 14 | VFB_jrchjzxy | LNd_R - 5813069648 | 29 | VFB_jrchjzxw | LNd_R - 5813056917 | LNd neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 5813056917 |
| 10 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | 25 | VFB_jrchjzxx | LNd_R - 5813021192 | s-LNv neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 511051477 | neuprint_JRC_Hemibrain_1point1 | 5813021192 |
| 8 | VFB_jrchjtf3 | DN1pA_R - 387166379 | 25 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1p neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 387166379 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 4 | VFB_jrchjtf0 | DN1pA_R - 324846570 | 25 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1p neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 324846570 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 12 | VFB_jrchjtez | DN1pA_R - 5813010153 | 25 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1p neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 5813010153 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 15 | VFB_jrchjtf2 | DN1pA_R - 387944118 | 22 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | DN1p neuron | s-LNv neuron | neuprint_JRC_Hemibrain_1point1 | 387944118 | neuprint_JRC_Hemibrain_1point1 | 511051477 |
| 30 | VFB_jrchjzxy | LNd_R - 5813069648 | 21 | VFB_jrchjzxx | LNd_R - 5813021192 | LNd neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 5813021192 |
| 2 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | 17 | VFB_jrchjzxw | LNd_R - 5813056917 | s-LNv neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 511051477 | neuprint_JRC_Hemibrain_1point1 | 5813056917 |
| 13 | VFB_jrchjtez | DN1pA_R - 5813010153 | 15 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 5813010153 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 24 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | 14 | VFB_jrchjtf3 | DN1pA_R - 387166379 | s-LNv neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 511051477 | neuprint_JRC_Hemibrain_1point1 | 387166379 |
| 25 | VFB_jrchjzxy | LNd_R - 5813069648 | 14 | VFB_jrchjtf1 | DN1pA_R - 325529237 | LNd neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 325529237 |
| 0 | VFB_jrchjtf0 | DN1pA_R - 324846570 | 13 | VFB_jrchjzxx | LNd_R - 5813021192 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 324846570 | neuprint_JRC_Hemibrain_1point1 | 5813021192 |
| 17 | VFB_jrchjzxy | LNd_R - 5813069648 | 13 | VFB_jrchjtf3 | DN1pA_R - 387166379 | LNd neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 387166379 |
| 9 | VFB_jrchk8e0 | 5th s-LNv - 511051477 | 12 | VFB_jrchjtf1 | DN1pA_R - 325529237 | s-LNv neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 511051477 | neuprint_JRC_Hemibrain_1point1 | 325529237 |
| 19 | VFB_jrchjtf5 | DN1pB_R - 5813071319 | 11 | VFB_jrchjzxy | LNd_R - 5813069648 | DN1p neuron | LNd neuron | neuprint_JRC_Hemibrain_1point1 | 5813071319 | neuprint_JRC_Hemibrain_1point1 | 5813069648 |
| 31 | VFB_jrchjtey | DN1a_R - 5813022274 | 11 | VFB_jrchjtex | DN1a_R - 264083994 | DN1a neuron | DN1a neuron | neuprint_JRC_Hemibrain_1point1 | 5813022274 | neuprint_JRC_Hemibrain_1point1 | 264083994 |
| 18 | VFB_jrchjzxy | LNd_R - 5813069648 | 10 | VFB_jrchjtez | DN1pA_R - 5813010153 | LNd neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 5813010153 |
| 27 | VFB_jrchjtex | DN1a_R - 264083994 | 10 | VFB_jrchjtey | DN1a_R - 5813022274 | DN1a neuron | DN1a neuron | neuprint_JRC_Hemibrain_1point1 | 264083994 | neuprint_JRC_Hemibrain_1point1 | 5813022274 |
| 6 | VFB_jrchjzxy | LNd_R - 5813069648 | 10 | VFB_jrchjtf2 | DN1pA_R - 387944118 | LNd neuron | DN1p neuron | neuprint_JRC_Hemibrain_1point1 | 5813069648 | neuprint_JRC_Hemibrain_1point1 | 387944118 |
# Get connections between visual projection neurons and descending neurons
vc.get_connected_neurons_by_type(upstream_type='visual projection neuron',
downstream_type='adult descending neuron',
weight=10).sort_values('weight', ascending=False)
| upstream_neuron_id | upstream_neuron_name | weight | downstream_neuron_id | downstream_neuron_name | upstream_class | downstream_class | up_data_source | up_accession | down_source | down_accession | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 966 | VFB_jrchk09j | LT51(aSP29a)_R - 1282348408 | 319 | VFB_jrchjtfr | DNb01_R - 1566597156 | lobula tangential neuron | descending neuron of the anterior ventral brai... | neuprint_JRC_Hemibrain_1point1 | 1282348408 | neuprint_JRC_Hemibrain_1point1 | 1566597156 |
| 682 | VFB_jrchk09c | LT51_R - 1444351896 | 221 | VFB_jrchjtfg | DNa03_R - 1139909038 | lobula tangential neuron | descending neuron of the anterior dorsal brain... | neuprint_JRC_Hemibrain_1point1 | 1444351896 | neuprint_JRC_Hemibrain_1point1 | 1139909038 |
| 559 | VFB_jrchk09j | LT51(aSP29a)_R - 1282348408 | 169 | VFB_jrchjthb | DNp18_R - 5813068840 | lobula tangential neuron | descending neuron of the posterior brain DNp18 | neuprint_JRC_Hemibrain_1point1 | 1282348408 | neuprint_JRC_Hemibrain_1point1 | 5813068840 |
| 744 | VFB_jrchk09i | LT51_R - 1597493900 | 141 | VFB_jrchjtfg | DNa03_R - 1139909038 | lobula tangential neuron | descending neuron of the anterior dorsal brain... | neuprint_JRC_Hemibrain_1point1 | 1597493900 | neuprint_JRC_Hemibrain_1point1 | 1139909038 |
| 961 | VFB_jrchk09l | LT51(aSP29a)_R - 1261229600 | 104 | VFB_jrchjtfr | DNb01_R - 1566597156 | lobula tangential neuron | descending neuron of the anterior ventral brai... | neuprint_JRC_Hemibrain_1point1 | 1261229600 | neuprint_JRC_Hemibrain_1point1 | 1566597156 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 284 | VFB_jrchjy92 | LC4 - 5901215446 | 10 | VFB_jrchjtg8 | DNp05_R - 1466998977 | lobula columnar neuron LC4 | descending neuron of the posterior brain DNp05 | neuprint_JRC_Hemibrain_1point1 | 5901215446 | neuprint_JRC_Hemibrain_1point1 | 1466998977 |
| 710 | VFB_jrchjznt | LLPC1 - 5812996978 | 10 | VFB_jrchjthl | DNp26_R - 1100404634 | lobula complex columnar neuron | descending neuron of the posterior brain DNp26 | neuprint_JRC_Hemibrain_1point1 | 5812996978 | neuprint_JRC_Hemibrain_1point1 | 1100404634 |
| 713 | VFB_jrchjzx3 | LLPC3 - 1873811611 | 10 | VFB_jrchjthl | DNp26_R - 1100404634 | lobula complex columnar neuron | descending neuron of the posterior brain DNp26 | neuprint_JRC_Hemibrain_1point1 | 1873811611 | neuprint_JRC_Hemibrain_1point1 | 1100404634 |
| 953 | VFB_jrchjzrz | LLPC2c - 5812997067 | 10 | VFB_jrchjtfh | DNa04_R - 1262014782 | lobula complex columnar neuron | descending neuron of the anterior dorsal brain... | neuprint_JRC_Hemibrain_1point1 | 5812997067 | neuprint_JRC_Hemibrain_1point1 | 1262014782 |
| 0 | VFB_jrchk04n | LPLC2_R - 5812987894 | 10 | VFB_jrchjtg7 | DNp04_R - 1405231475 | lobula complex columnar neuron LPLC2 | descending neuron of the posterior brain DNp04 | neuprint_JRC_Hemibrain_1point1 | 5812987894 | neuprint_JRC_Hemibrain_1point1 | 1405231475 |
1085 rows × 11 columns
Get neurons downstream/downstream of a specific neuron
Starting from an individual neuron, in this case an individual neuron from hemibrain, find all neurons upstream or downstream, restricting to edges >= some specified wieght (synaptic connections) and optionally limiting the output by neuron type/class.
# Starting point - let's take one of the individual descending neurons from
# the previous exploratory queries and look for other inputs
vc.get_neurons_upstream_of('DNp04_R - 1405231475', weight = 20)
| query_neuron_id | query_neuron_name | weight | target_neuron_id | target_neuron_name | |
|---|---|---|---|---|---|
| 0 | VFB_jrchk4u3 | PVLP100_R - 1375845363 | 44 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 1 | VFB_jrchjy8u | LC4 - 1158864995 | 50 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 2 | VFB_jrchjy9f | LC4 - 5813055129 | 46 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 3 | VFB_jrchjyal | LC4 - 1907571222 | 78 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 4 | VFB_jrchjy9t | LC4 - 2215161310 | 86 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| ... | ... | ... | ... | ... | ... |
| 82 | VFB_jrchjyak | LC4 - 1907933561 | 82 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 83 | VFB_jrchjy9d | LC4 - 1627117134 | 46 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 84 | VFB_jrchjya7 | LC4 - 1876471221 | 58 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 85 | VFB_jrchk4tm | PVLP094_R - 1503733177 | 26 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 86 | VFB_jrchjya0 | LC4 - 5812998136 | 92 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
87 rows × 5 columns
Let’s try a similar query from one of the LC4 neurons to see what other descending neuron types are downstream of it.
vc.get_neurons_downstream_of('LC4 - 1249932198', classification="'adult descending neuron'", weight = 20)
Running query: FBbt:00047511
Query URL: http://owl.virtualflybrain.org/kbs/vfb/instances?object=FBbt%3A00047511&prefixes=%7B%22FBbt%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FFBbt_%22%2C+%22RO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FRO_%22%2C+%22BFO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FBFO_%22%7D&direct=False
Query results: 139
| query_neuron_id | query_neuron_name | weight | target_neuron_id | target_neuron_name | |
|---|---|---|---|---|---|
| 0 | VFB_jrchjy8y | LC4 - 1249932198 | 78 | VFB_jrchjtg7 | DNp04_R - 1405231475 |
| 1 | VFB_jrchjy8y | LC4 - 1249932198 | 30 | VFB_jrchjup1 | Giant Fiber_R - 2307027729 |
| 2 | VFB_jrchjy8y | LC4 - 1249932198 | 65 | VFB_jrchjtgf | DNp11_R - 1281324958 |
More sophisticated connectomics queries require direct queries of connectomics DB APIs
CATMAID connectivity queries
Broadly speaking, using pymaid to query CATMAID servers lets you fetch connectivity data as either lists of up- and downstream partners or as whole adjacency matrices.
These examples use the VFB FAFB server.
# Using VFB to get neurons by type
DA1 = vc.get_instances("'adult antennal lobe projection neuron DA1'", summary=True)
DA1_tab = pd.DataFrame.from_records(DA1)
len(DA1_tab)
DA1_tab
Running query: FBbt:00048096
Query URL: http://owl.virtualflybrain.org/kbs/vfb/instances?object=FBbt%3A00048096&prefixes=%7B%22FBbt%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FFBbt_%22%2C+%22RO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FRO_%22%2C+%22BFO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FBFO_%22%7D&direct=False
Query results: 31
| label | symbol | id | tags | parents_label | parents_id | data_source | accession | templates | dataset | license | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Uniglomerular mALT DA1 lPN#L3 (FAFB:2345089) | VFB_0010122p | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2345089 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 1 | ALv1_P02(DA1)_0_2018U | VFB_00102763 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | Yu2013 | https://creativecommons.org/licenses/by-nc-sa/... | ||||
| 2 | Uniglomerular mALT DA1 lPN#R3 (FAFB:61221) | VFB_00101204 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 61221 | adult brain template JFRC2|JRC2018Unisex | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 3 | Uniglomerular mALT DA1 lPN#L1 (FAFB:4207871) | VFB_0010126e | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 4207871 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 4 | ALv1_P02(DA1)_0_2018U | VFB_00103943 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | Yu2013 | https://creativecommons.org/licenses/by-nc-sa/... | ||||
| 5 | Uniglomerular mALT DA1 lPN#R1 (FAFB:57323) | VFB_00101201 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 57323 | JRC2018Unisex|adult brain template JFRC2 | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 6 | DA1_lPN_R - 1734350908 | VFB_jrchjtdb | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 1734350908 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 7 | Uniglomerular mALT DA1 lPN#R5 (FAFB:2863104) | VFB_0010124e | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2863104 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 8 | Uniglomerular mALT DA1 lPN#R7 (FAFB:57353) | VFB_00101202 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 57353 | JRC2018Unisex|adult brain template JFRC2 | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 9 | ALv1_P02(DA1)_0_2018U | VFB_00102297 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | JRC2018Unisex | Lee2020 | https://creativecommons.org/licenses/by/4.0/le... | |||
| 10 | ALv1_P02(DA1)_0_2018U | VFB_00104629 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | JRC2018Unisex | Yu2013 | https://creativecommons.org/licenses/by-nc-sa/... | |||
| 11 | ALv1_P02(DA1)_0_2018U | VFB_00102294 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | Yu2013 | https://creativecommons.org/licenses/by-nc-sa/... | ||||
| 12 | Uniglomerular mlALT DA1 vPN#R1 (FAFB:1811442) | VFB_0010121x | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | catmaid_fafb | 1811442 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 13 | Uniglomerular mlALT DA1 vPN#L1 (FAFB:2334841) | VFB_0010122m | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | catmaid_fafb | 2334841 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 14 | Uniglomerular mALT DA1 lPN#L5 (FAFB:2380564) | VFB_0010122z | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2380564 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 15 | DA1_lPN_R - 754538881 | VFB_jrchjtdg | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 754538881 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 16 | Uniglomerular mALT DA1 lPN#R6 (FAFB:27295) | VFB_00101199 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 27295 | JRC2018Unisex|adult brain template JFRC2 | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 17 | Uniglomerular mALT DA1 lPN#R8 (FAFB:57381) | VFB_00101203 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 57381 | adult brain template JFRC2|JRC2018Unisex | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 18 | DA1_lPN_R - 1765040289 | VFB_jrchjtdc | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 1765040289 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 19 | Uniglomerular mALT DA1 lPN#R2 (FAFB:57311) | VFB_00101200 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 57311 | adult brain template JFRC2|JRC2018Unisex | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 20 | DA1_lPN_R - 5813039315 | VFB_jrchjtdd | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 5813039315 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 21 | Uniglomerular mALT DA1 lPN#L6 (FAFB:2381753) | VFB_0010123b | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2381753 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 22 | DA1_lPN_R - 754534424 | VFB_jrchjtde | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 754534424 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 23 | DA1_lPN_R - 1734350788 | VFB_jrchjtdf | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 1734350788 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 24 | ALv1_P02(DA1)_0_2018U | VFB_00104624 | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | Yu2013 | https://creativecommons.org/licenses/by-nc-sa/... | ||||
| 25 | Uniglomerular mALT DA1 lPN#L2 (FAFB:2319457) | VFB_0010122k | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2319457 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 26 | Uniglomerular mALT DA1 lPN#R4 (FAFB:755022) | VFB_00101205 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 755022 | adult brain template JFRC2|JRC2018Unisex | Zheng2018 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 27 | Uniglomerular mALT DA1 lPN#L7 (FAFB:3239781) | VFB_0010124l | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 3239781 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 28 | Uniglomerular mALT DA1 lPN#L4 (FAFB:2379517) | VFB_0010122y | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | catmaid_fafb | 2379517 | JRC2018Unisex | BatesSchlegel2020 | https://creativecommons.org/licenses/by-sa/4.0... | |
| 29 | DA1_vPN_R - 733316908 | VFB_jrchjtdh | Entity|GABAergic|Adult|Anatomy|has_image|Cell|... | adult antennal lobe projection neuron DA1 vPN | FBbt_00067372 | neuprint_JRC_Hemibrain_1point1 | 733316908 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 30 | DA1_lPN_R - 722817260 | VFB_jrchjtda | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DA1 lPN | FBbt_00067363 | neuprint_JRC_Hemibrain_1point1 | 722817260 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... |
# Map neurons to CATMAID Skeleton IDs (skids)
da1_skids = vc.neo_query_wrapper.vfb_id_2_xrefs(DA1_tab['id'], db='catmaid_fafb', reverse_return=True)
da1_skids
{'2863104': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010124e'}],
'57353': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101202'}],
'2381753': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010123b'}],
'1811442': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010121x'}],
'57323': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101201'}],
'57311': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101200'}],
'2345089': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010122p'}],
'27295': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101199'}],
'2379517': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010122y'}],
'2380564': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010122z'}],
'755022': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101205'}],
'61221': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101204'}],
'57381': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_00101203'}],
'3239781': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010124l'}],
'4207871': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010126e'}],
'2334841': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010122m'}],
'2319457': [{'db': 'catmaid_fafb', 'vfb_id': 'VFB_0010122k'}]}
# Map neurons to CATMAID Skeleton IDs (skids)
da1_skids = vc.neo_query_wrapper.vfb_id_2_xrefs(DA1_tab['id'], db='catmaid_fafb', reverse_return=True)
da1_skids_int = list(map(int, da1_skids))
da1_skids_int
[2863104,
57353,
2381753,
1811442,
57323,
57311,
2345089,
27295,
2379517,
2380564,
755022,
61221,
57381,
3239781,
4207871,
2334841,
2319457]
Generate a connectivity table for neurons downstream of DA1 neurons
da1 = pymaid.get_neurons(da1_skids_int)
da1_ds = pymaid.get_partners(da1_skids_int,
threshold=3, # anything with >= 3 synapses
directions=['outgoing'] # downstream partners only
)
# Result is a pandas DataFrame
da1_ds.head()
INFO : Fetching connectivity table for 17 neurons (pymaid)
INFO : Done. Found 0 pre-, 270 postsynaptic and 0 gap junction-connected neurons (pymaid)
| neuron_name | skeleton_id | num_nodes | relation | 2863104 | 57353 | 2381753 | 1811442 | 57323 | 57311 | ... | 2379517 | 2380564 | 755022 | 61221 | 57381 | 3239781 | 4207871 | 2334841 | 2319457 | total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Uniglomerular mlALT DA1 vPN mlALTed Milk 18114... | 1811442 | 11769 | downstream | 30 | 3 | 0 | 0 | 20 | 21 | ... | 0 | 0 | 32 | 26 | 4 | 0 | 0 | 0 | 0 | 151.0 |
| 1 | Uniglomerular mlALT DA1 vPN mlALTed Milk 23348... | 2334841 | 6362 | downstream | 0 | 0 | 32 | 0 | 0 | 0 | ... | 28 | 0 | 0 | 0 | 0 | 26 | 17 | 0 | 22 | 139.0 |
| 2 | LHAV4a4#1 1911125 FML PS RJVR | 1911124 | 6969 | downstream | 23 | 6 | 0 | 0 | 15 | 19 | ... | 0 | 0 | 19 | 13 | 9 | 0 | 0 | 0 | 0 | 109.0 |
| 3 | LHAV2a3#1 1870231 RJVR AJES PS | 1870230 | 14820 | downstream | 5 | 23 | 0 | 1 | 7 | 5 | ... | 0 | 0 | 19 | 7 | 28 | 0 | 0 | 0 | 0 | 105.0 |
| 4 | LHAV4c1#1 488056 downstream DA1 GSXEJ | 488055 | 12137 | downstream | 15 | 3 | 0 | 0 | 11 | 17 | ... | 0 | 0 | 15 | 15 | 0 | 0 | 0 | 0 | 0 | 92.0 |
5 rows × 22 columns
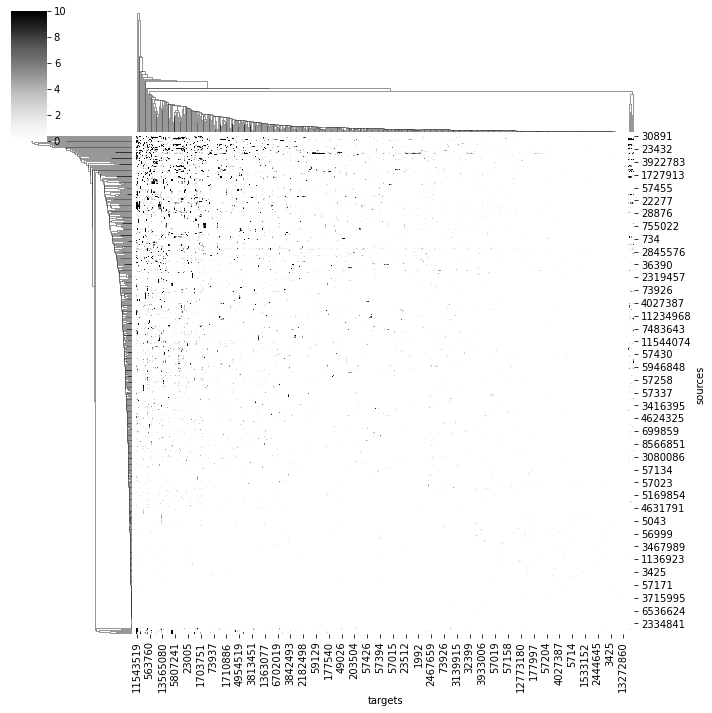
# Get an adjacency matrix between all Bates, Schlegel et al. neurons
bates = pymaid.find_neurons(annotations='Paper: Bates and Schlegel et al 2020')
adj = pymaid.adjacency_matrix(bates)
adj.head()
INFO : Found 583 neurons matching the search parameters (pymaid)
| targets | 2863104 | 57349 | 57353 | 16 | 57361 | 15738898 | 57365 | 4182038 | 3813399 | 11524119 | ... | 57323 | 4624362 | 1853423 | 2842610 | 57333 | 4624374 | 3080183 | 57337 | 4624378 | 57341 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sources | |||||||||||||||||||||
| 2863104 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 2.0 | 0.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 57349 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 57353 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 57361 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
5 rows × 583 columns
import seaborn as sns
ax = sns.clustermap(adj, vmax=10, cmap='Greys')
/shared-libs/python3.7/py/lib/python3.7/site-packages/seaborn/matrix.py:649: UserWarning:
Clustering large matrix with scipy. Installing `fastcluster` may give better performance.
# Axo-axonic connections between two different types of DA1 PNs
cn = pymaid.get_connectors_between(2863104, 1811442)
cn.head()
| connector_id | connector_loc | node1_id | source_neuron | confidence1 | creator1 | node1_loc | node2_id | target_neuron | confidence2 | creator2 | node2_loc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6736296 | [359448.44, 159319.03, 150560.0] | 3163408 | 2863104 | 5 | NaN | [359487.3, 159145.66, 150600.0] | 6736298 | 1811442 | 5 | NaN | [359611.9, 159541.48, 150560.0] |
| 1 | 6795172 | [356041.88, 149555.53, 147920.0] | 6795195 | 2863104 | 5 | NaN | [354724.44, 149284.1, 147920.0] | 6795153 | 1811442 | 5 | NaN | [356366.16, 149854.86, 147920.0] |
| 2 | 6795291 | [355189.5, 150232.48, 148240.0] | 6795293 | 2863104 | 5 | NaN | [354595.62, 149464.8, 148240.0] | 6795214 | 1811442 | 5 | NaN | [355472.28, 150294.75, 148160.0] |
| 3 | 6795747 | [355030.4, 154047.86, 145800.0] | 6795749 | 2863104 | 5 | NaN | [355045.38, 154180.1, 145800.0] | 6795745 | 1811442 | 5 | NaN | [355024.44, 153945.73, 145760.0] |
| 4 | 6797452 | [353221.4, 148570.9, 147320.0] | 6797456 | 2863104 | 5 | NaN | [354213.9, 148397.44, 147320.0] | 6797437 | 1811442 | 5 | NaN | [353447.6, 148704.88, 147560.0] |
import numpy as np
points = np.vstack(cn.connector_loc)
navis.plot3d([da1.idx[[1811442, 57353]], # plot the two neurons
points], # plot the points of synaptic contacts as scatter
scatter_kws=dict(name="synaptic contacts")
)
#neu_da1 = neu.fetch_skeletons(neu.NeuronCriteria(type='DA1_lPN.*', regex=True))
neu_da1
| type | name | id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | navis.TreeNeuron | DA1_lPN_R | 722817260 | 4336 | 3132 | 635 | 658 | 274910.568784 | NaN | 8 nanometer |
| 1 | navis.TreeNeuron | DA1_lPN_R | 754538881 | 4890 | 2943 | 626 | 642 | 291434.992623 | 860.0 | 8 nanometer |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5 | navis.TreeNeuron | DA1_lPN_R | 1765040289 | 5380 | 3099 | 624 | 642 | 324249.403013 | 2467.0 | 8 nanometer |
| 6 | navis.TreeNeuron | DA1_lPN_R | 5813039315 | 4300 | 2954 | 625 | 643 | 273403.049029 | NaN | 8 nanometer |
split = navis.split_axon_dendrite(neu_da1[0])
split
| type | name | id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | compartment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | navis.TreeNeuron | DA1_lPN_R | 722817260 | 0 | 0 | 0 | 0 | 0.000000 | None | 8 nanometer | cellbodyfiber |
| 1 | navis.TreeNeuron | DA1_lPN_R | 722817260 | 3575 | 2505 | 566 | 590 | 188806.744034 | None | 8 nanometer | dendrite |
| 2 | navis.TreeNeuron | DA1_lPN_R | 722817260 | 193 | 14 | 0 | 1 | 30601.319501 | None | 8 nanometer | linker |
| 3 | navis.TreeNeuron | DA1_lPN_R | 722817260 | 474 | 612 | 60 | 61 | 48872.448763 | None | 8 nanometer | axon |
navis.plot3d(split, connectors=True)
WARNING : Skipping empty neuron: DA1_lPN_R (722817260) (navis)
cn = neu.fetch_synapse_connections(source_criteria=neu.NeuronCriteria(type='DA1_lPN.*', regex=True),
target_criteria=neu.NeuronCriteria(type='DA1_vPN.*', regex=True))
cn.head()
| bodyId_pre | bodyId_post | roi_pre | roi_post | x_pre | y_pre | z_pre | x_post | y_post | z_post | confidence_pre | confidence_post | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 754534424 | 733316908 | LH(R) | LH(R) | 5015 | 22921 | 15043 | 5033 | 22941 | 15044 | 0.972 | 0.991824 |
| 1 | 754534424 | 733316908 | AL(R) | AL(R) | 14588 | 35361 | 26453 | 14582 | 35377 | 26444 | 0.991 | 0.932866 |
| 2 | 754534424 | 733316908 | LH(R) | LH(R) | 3826 | 22574 | 14689 | 3842 | 22576 | 14683 | 0.982 | 0.896622 |
| 3 | 754534424 | 733316908 | LH(R) | LH(R) | 4924 | 23750 | 14333 | 4927 | 23768 | 14324 | 0.993 | 0.961103 |
| 4 | 754534424 | 733316908 | LH(R) | LH(R) | 5017 | 23642 | 14531 | 4997 | 23622 | 14518 | 0.904 | 0.661779 |
cn.groupby(['bodyId_pre', 'bodyId_post', 'roi_pre']).size()
bodyId_pre bodyId_post roi_pre
722817260 733316908 AL(R) 26
LH(R) 28
754534424 733316908 AL(R) 13
LH(R) 31
754538881 733316908 AL(R) 33
LH(R) 5
1734350788 733316908 AL(R) 21
LH(R) 38
1734350908 733316908 AL(R) 24
LH(R) 37
1765040289 733316908 AL(R) 27
LH(R) 2
5813039315 733316908 AL(R) 12
LH(R) 33
dtype: int64
neuprint connectivity queries
To programmatically interface with neuprint, we will use neuprint-python (link). It requires an API token which you can get via the website and is bound to the Google account that you use to log into neuprint. For this workshop we provide such a token as environment variable but you will need to start using your own token after the workshop is over.
These examples use the hemibrain v1.1 dataset.
neuprint-python
First we have to initialize the connection.
import neuprint as neu
client = neu.Client('https://neuprint.janelia.org',
dataset='hemibrain:v1.1',
token='eyJhbGciOiJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJlbWFpbCI6InZmYndvcmtzaG9wLm5ldXJvZmx5MjAyMEBnbWFpbC5jb20iLCJsZXZlbCI6Im5vYXV0aCIsImltYWdlLXVybCI6Imh0dHBzOi8vbGg2Lmdvb2dsZXVzZXJjb250ZW50LmNvbS8tWXFDN21NRXd3TlEvQUFBQUFBQUFBQUkvQUFBQUFBQUFBQUEvQU1adXVjbU5zaXhXZDRhM0VyTTQ0ODBMa2IzNDdvUlpfUS9zOTYtYy9waG90by5qcGc_c3o9NTA_c3o9NTAiLCJleHAiOjE3OTQwOTE4ODd9.ceg4mrj2o-aOhK0NHNGmBacg8R34PBPoLBwhCo4uOCQ')
Most functions in neuprint-python accept neu.NeuronCriteria which is effectively a filter for body IDs, types, cellbody fibers (~lineages), etc:
help(neu.NeuronCriteria)
Help on class NeuronCriteria in module neuprint.neuroncriteria:
class NeuronCriteria(builtins.object)
| NeuronCriteria(matchvar='n', *, bodyId=None, instance=None, type=None, regex=False, cellBodyFiber=None, status=None, cropped=None, min_pre=0, min_post=0, rois=None, inputRois=None, outputRois=None, min_roi_inputs=1, min_roi_outputs=1, label=None, roi_req='all', client=None)
|
| Specifies which fields to filter by when searching for a Neuron (or Segment).
| This class does not send queries itself, but you use it to specify search
| criteria for various query functions.
|
| Note:
| For simple queries involving only particular bodyId(s) or type(s)/instance(s),
| you can usually just pass the ``bodyId`` or ``type`` to the query function,
| without constructing a full ``NeuronCriteria``.
|
| .. code-block:: python
|
| from neuprint import fetch_neurons, NeuronCriteria as NC
|
| # Equivalent
| neuron_df, conn_df = fetch_neurons(NC(bodyId=329566174))
| neuron_df, conn_df = fetch_neurons(329566174)
|
| # Equivalent
| # (Criteria is satisfied if either type or instance matches.)
| neuron_df, conn_df = fetch_neurons(NC(type="OA-VPM3", instance="OA-VPM3"))
| neuron_df, conn_df = fetch_neurons("OA-VPM3")
|
| Methods defined here:
|
| __eq__(self, value)
| Implement comparison between criteria.
| Note: 'matchvar' is not considered during the comparison.
|
| __init__(self, matchvar='n', *, bodyId=None, instance=None, type=None, regex=False, cellBodyFiber=None, status=None, cropped=None, min_pre=0, min_post=0, rois=None, inputRois=None, outputRois=None, min_roi_inputs=1, min_roi_outputs=1, label=None, roi_req='all', client=None)
| Except for ``matchvar``, all parameters must be passed as keyword arguments.
|
| .. note::
|
| **Options for specifying ROI criteria**
|
| The ``rois`` argument merely matches neurons that intersect the given ROIs at all
| (without distinguishing between inputs and outputs).
|
| The ``inputRois`` and ``outputRois`` arguments allow you to put requirements
| on whether or not neurons have inputs or outputs in the listed ROIs.
| It results a more expensive query, but its more powerful.
| It also enables you to require a minimum number of connections in the given
| ``inputRois`` or ``outputRois`` using the ``min_roi_inputs`` and ``min_roi_outputs``
| criteria.
|
| In either case, use use ``roi_req`` to specify whether a neuron must match just
| one (``any``) of the listed ROIs, or ``all`` of them.
|
| Args:
| matchvar (str):
| An arbitrary cypher variable name to use when this
| ``NeuronCriteria`` is used to construct cypher queries.
| To help catch errors (such as accidentally passing a ``type`` or
| ``instance`` name in the wrong argument position), we require that
| ``matchvar`` begin with a lowercase letter.
|
| bodyId (int or list of ints):
| List of bodyId values.
|
| instance (str or list of str):
| If ``regex=True``, then the instance will be matched as a regular expression.
| Otherwise, only exact matches are found. To search for neurons with no instance
| at all, use ``instance=[None]``. If both ``type`` and ``instance`` criteria are
| supplied, any neuron that matches EITHER criteria will match the overall criteria.
|
| type (str or list of str):
| If ``regex=True``, then the type will be matched as a regular expression.
| Otherwise, only exact matches are found. To search for neurons with no type
| at all, use ``type=[None]``. If both ``type`` and ``instance`` criteria are
| supplied, any neuron that matches EITHER criteria will match the overall criteria.
|
| regex (bool):
| If ``True``, the ``instance`` and ``type`` arguments will be interpreted as
| regular expressions, rather than exact match strings.
|
| cellBodyFiber (str or list of str):
| Matches for the neuron ``cellBodyFiber`` field. To search for neurons
| with no CBF at all, use ``cellBodyFiber=[None]``.
|
| status (str or list of str):
| Matches for the neuron ``status`` field. To search for neurons with no status
| at all, use ``status=[None]``.
|
| cropped (bool):
| If given, restrict results to neurons that are cropped or not.
|
| min_pre (int):
| Exclude neurons that don't have at least this many t-bars (outputs) overall,
| regardless of how many t-bars exist in any particular ROI.
|
| min_post (int):
| Exclude neurons that don't have at least this many PSDs (inputs) overall,
| regardless of how many PSDs exist in any particular ROI.
|
| rois (str or list of str):
| ROIs that merely intersect the neuron, without specifying whether
| they're intersected by input or output synapses.
| If not provided, will be auto-set from ``inputRois`` and ``outputRois``.
|
| inputRois (str or list of str):
| Only Neurons which have inputs in EVERY one of the given ROIs will be matched.
| ``regex`` does not apply to this parameter.
|
| outputRois (str or list of str):
| Only Neurons which have outputs in EVERY one of the given ROIs will be matched.
| ``regex`` does not apply to this parameter.
|
| min_roi_inputs (int):
| How many input (post) synapses a neuron must have in each ROI to satisfy the
| ``inputRois`` criteria. Can only be used if you provided ``inputRois``.
|
| min_roi_outputs (int):
| How many output (pre) synapses a neuron must have in each ROI to satisfy the
| ``outputRois`` criteria. Can only be used if you provided ``outputRois``.
|
| roi_req (Either ``'any'`` or ``'all'``):
| Whether a neuron must intersect all of the listed input/output ROIs, or any of the listed input/output ROIs.
| When using 'any', each neuron must still match at least one input AND at least one output ROI.
|
| label (Either ``'Neuron'`` or ``'Segment'``):
| Which node label to match with.
| (In neuprint, all ``Neuron`` nodes are also ``Segment`` nodes.)
| By default, ``'Neuron'`` is used, unless you provided a non-empty ``bodyId`` list.
| In that case, ``'Segment'`` is the default. (It's assumed you're really interested
| in the bodies you explicitly listed, whether or not they have the ``'Neuron'`` label.)
|
| client (:py:class:`neuprint.client.Client`):
| Used to validate ROI names.
| If not provided, the global default ``Client`` will be used.
|
| __repr__(self)
| Return repr(self).
|
| all_conditions(self, *vars, prefix=0, comments=True)
|
| basic_conditions(self, prefix=0, comments=True)
| Construct a WHERE clause based on the basic conditions
| in this criteria (i.e. everything except for the "directed ROI" conditions.)
|
| basic_exprs(self)
| Return the list of expressions that correspond
| to the members in this NeuronCriteria object.
| They're intended be combined (via 'AND') in
| the WHERE clause of a cypher query.
|
| bodyId_expr(self)
|
| cbf_expr(self)
|
| cropped_expr(self)
|
| directed_rois_condition(self, *vars, prefix=0, comments=True)
| Construct the ```WITH...WHERE``` statements that apply the "directed ROI"
| conditions specified by this criteria's ``inputRois`` and ``outputRois``
| members.
|
| These conditions are expensive to evaluate, so it's usually a good
| idea to position them LAST in your cypher query, once the result set
| has already been narrowed down by eariler filters.
|
| global_vars(self)
|
| global_with(self, *vars, prefix=0)
|
| instance_expr(self)
|
| post_expr(self)
|
| pre_expr(self)
|
| rois_expr(self)
|
| status_expr(self)
|
| type_expr(self)
|
| typeinst_expr(self)
| Unlike all other fields, type and instance OR'd together.
| Either match satisfies the criteria.
|
| ----------------------------------------------------------------------
| Class methods defined here:
|
| combined_conditions(neuron_conditions, vars=[], prefix=0, comments=True) from builtins.type
| Combine the conditions from multiple NeuronCriteria into a single string,
| putting the "cheap" conditions first and the "expensive" conditions last.
| (That is, basic conditions first and the directed ROI conditions last.)
|
| combined_global_with(neuron_conditions, vars=[], prefix=0) from builtins.type
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| MAX_LITERAL_LENGTH = 3
|
| __hash__ = None
Fetching neurons
Let’s say we want to find all antennnal lobe projection neurons (PNs). Their type nomenclature adheres to {glomerulus}_{lineage}PN (e.g. DA1_lPN)for uniglomerular PNs and a M_{lineage}PN{tract}{type} (e.g. M_vPNml50 = “multiglomerular ventral lineage PN mediolateral tract type 50) for multiglomerular PNs.
To get them all, we need to use regex patterns (see this cheatsheet):
# Define the filter criteria
nc = neu.NeuronCriteria(type='.*?_.*?PN.*?', regex=True)
# Get general info for these neurons
pns, roi_info = neu.fetch_neurons(nc)
print(f'{pns.shape[0]} PNs found.')
pns.head()
337 PNs found.
| bodyId | instance | type | pre | post | size | status | cropped | statusLabel | cellBodyFiber | somaRadius | somaLocation | inputRois | outputRois | roiInfo | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 294792184 | M_vPNml53_R | M_vPNml53 | 92 | 344 | 420662445 | Traced | False | Roughly traced | AVM04 | 336.5 | [18923, 34319, 35424] | [AL(R), AL-D(R), AL-DA2(R), AL-DA4m(R), AL-DC1... | [AL(R), AL-DC1(R), LH(R), PLP(R), SIP(R), SLP(... | {'SNP(R)': {'pre': 70, 'post': 155, 'downstrea... |
| 1 | 329599710 | M_lvPNm32_R | M_lvPNm32 | 247 | 285 | 343478957 | Traced | False | Roughly traced | AVM06 | NaN | None | [AL(R), AL-DC4(R), AL-DL2v(R), AL-DM1(R), AL-D... | [AL(R), AL-DL2v(R), AL-DM1(R), AL-DM4(R), AL-D... | {'SNP(R)': {'pre': 180, 'post': 93, 'downstrea... |
| 2 | 417199910 | M_lvPNm36_R | M_lvPNm36 | 162 | 347 | 387058559 | Traced | False | Roughly traced | AVM06 | 351.5 | [13823, 33925, 34176] | [AL(R), AL-DL5(R), AL-DM4(R), AL-DP1m(R), AL-V... | [AL(R), AL-DL5(R), AL-DM4(R), AL-VP1d(R), AL-V... | {'SNP(R)': {'pre': 156, 'post': 95, 'downstrea... |
| 3 | 480927537 | M_vPNml70_R | M_vPNml70 | 82 | 276 | 240153322 | Traced | False | Roughly traced | AVM04 | NaN | None | [AL(R), AL-DA2(R), AL-DA4l(R), AL-DA4m(R), AL-... | [LH(R), SLP(R), SNP(R)] | {'SNP(R)': {'pre': 15, 'post': 18, 'downstream... |
| 4 | 481268653 | M_vPNml89_R | M_vPNml89 | 146 | 58 | 265085609 | Traced | False | Roughly traced | AVM04 | NaN | None | [AL(R), AL-VC3l(R), AL-VC4(R), AL-VP1m(R), LH(... | [LH(R), SLP(R), SNP(R)] | {'SNP(R)': {'pre': 10, 'post': 2, 'downstream'... |
# Check that the regex did not have any accidental by-catch
pns['type'].unique()
array(['M_vPNml53', 'M_lvPNm32', 'M_lvPNm36', 'M_vPNml70', 'M_vPNml89',
'VP1l+_lvPN', 'M_vPNml69', 'DM1_lPN', 'DM4_vPN', 'M_vPNml79',
'VP4+_vPN', 'DA4l_adPN', 'M_vPNml87', 'DM4_adPN', 'M_vPNml83',
'VA5_lPN', 'DA4m_adPN', 'M_lvPNm24', 'M_vPNml85', 'VP1l+VP3_ilPN',
'M_vPNml77', 'M_vPNml84', 'VC1_lPN', 'M_lvPNm39', 'M_vPNml50',
'DM2_lPN', 'VC5_lvPN', 'M_vPNml88', 'M_vPNml58', 'VP4_vPN',
'DP1m_vPN', 'DP1m_adPN', 'DM5_lPN', 'VC5_adPN', 'M_vPNml80',
'M_lvPNm25', 'VC3m_lvPN', 'VP3+_vPN', 'VP1m+_lvPN', 'DA3_adPN',
'V_l2PN', 'M_vPNml56', 'VC3l_adPN', 'VM7v_adPN', 'DL5_adPN',
'VM4_adPN', 'VM2_adPN', 'M_lvPNm40', 'DC4_vPN', 'V_ilPN',
'M_vPNml74', 'Z_lvPNm1', 'DA1_lPN', 'DP1l_adPN', 'VM4_lvPN',
'M_vPNml71', 'DP1l_vPN', 'M_lvPNm41', 'M_spPN5t10', 'DA1_vPN',
'VC4_adPN', 'DM3_adPN', 'M_lvPNm45', 'VL1_vPN', 'M_lvPNm44',
'M_vPNml78', 'M_vPNml67', 'M_adPNm5', 'M_smPNm1', 'DM6_adPN',
'DL2d_adPN', 'M_adPNm6', 'M_adPNm8', 'M_lvPNm43', 'Z_vPNml1',
'M_vPNml59', 'DA2_lPN', 'M_lPNm11A', 'M_vPNml52', 'DL2d_vPN',
'VL2p_vPN', 'VA1d_adPN', 'M_lPNm11B', 'M_lvPNm48', 'M_lPNm11C',
'M_lvPNm42', 'VA1v_vPN', 'M_vPNml68', 'M_vPNml55', 'M_vPNml62',
'VL2a_vPN', 'M_vPNml60', 'M_vPNml65', 'VM5d_adPN', 'M_l2PNm16',
'M_vPNml61', 'M_vPNml57', 'M_vPNml64', 'M_lv2PN9t49',
'VP2+VC5_l2PN', 'M_spPN4t9', 'M_vPNml66', 'M_vPNml75', 'M_vPNml63',
'M_vPNml72', 'M_lvPNm38', 'D_adPN', 'M_vPNml76', 'M_vPNml54',
'DM3_vPN', 'M_vPNml86', 'DL3_lPN', 'VA4_lPN', 'VP1d_il2PN',
'DC1_adPN', 'M_l2PN3t18', 'M_lvPNm35', 'DL4_adPN', 'M_lvPNm28',
'M_lvPNm27', 'M_ilPNm90', 'M_l2PNl20', 'M_lvPNm29', 'VA7l_adPN',
'M_lPNm13', 'M_l2PNl21', 'DL1_adPN', 'M_imPNl92', 'M_vPNml73',
'M_ilPN8t91', 'M_l2PNm14', 'VP1d+VP4_l2PN1', 'M_lvPNm26',
'DL2v_adPN', 'VP3+VP1l_ivPN', 'M_lvPNm33', 'VA1v_adPN',
'VP3+_l2PN', 'M_l2PN10t19', 'VP4+VL1_l2PN', 'M_l2PNl22',
'M_l2PNm15', 'M_lPNm11D', 'MZ_lv2PN', 'DC2_adPN', 'M_lvPNm46',
'VC2_lPN', 'VM1_lPN', 'VM3_adPN', 'VM7d_adPN', 'M_lvPNm47',
'M_lPNm12', 'DC3_adPN', 'VP2+_adPN', 'VP1m+VP2_lvPN2',
'VP1m+VP2_lvPN1', 'VA6_adPN', 'VA7m_lPN', 'M_adPNm7', 'M_adPNm4',
'VA1d_vPN', 'VA3_adPN', 'VL1_ilPN', 'M_l2PNl23', 'M_lvPNm31',
'VP1m+VP5_ilPN', 'VL2p_adPN', 'MZ_lvPN', 'VP2_adPN', 'VA2_adPN',
'VM5v_adPN', 'VP5+VP2_l2PN', 'VP5+VP3_l2PN', 'VP5+_l2PN',
'M_vPNml51', 'M_smPN6t2', 'M_lvPNm37', 'M_vPNml82', 'M_adPNm3',
'VP1m_l2PN', 'DC4_adPN', 'VP5+Z_adPN', 'VL2a_adPN', 'VP2_l2PN',
'M_lvPNm34', 'VP2+Z_lvPN', 'M_lvPNm30', 'M_l2PNm17', 'M_vPNml81',
'VP1d+VP4_l2PN2'], dtype=object)
Find neuron types with VFB instead (this allows curated neuron classes to be used):
#This will get all ALPNs from ALL datasets on VFB
ALPNs = vc.get_instances("'adult antennal lobe projection neuron'", summary=True)
ALPNs = pd.DataFrame.from_records(ALPNs)
#Select only rows from Hemibrain1.1 dataset
ALPNs=ALPNs[ALPNs['data_source'].str.match('neuprint_JRC_Hemibrain_1point1')]
ALPNs
Running query: FBbt:00067123
Query URL: http://owl.virtualflybrain.org/kbs/vfb/instances?object=FBbt%3A00067123&prefixes=%7B%22FBbt%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FFBbt_%22%2C+%22RO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FRO_%22%2C+%22BFO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FBFO_%22%7D&direct=False
Query results: 1469
| label | symbol | id | tags | parents_label | parents_id | data_source | accession | templates | dataset | license | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | M_vPNml53_R - 294792184 | VFB_jrchk0y5 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00007414 | neuprint_JRC_Hemibrain_1point1 | 294792184 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 4 | DC2_adPN_R - 1828206773 | VFB_jrchjtdt | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron DC2 adPN | FBbt_00067354 | neuprint_JRC_Hemibrain_1point1 | 1828206773 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 8 | VM4_lvPN_R - 1850024931 | VFB_jrchk7ib | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron VM4 lvPN | FBbt_00049775 | neuprint_JRC_Hemibrain_1point1 | 1850024931 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 18 | M_vPNml81_R - 5813090752 | VFB_jrchk0zj | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00007414 | neuprint_JRC_Hemibrain_1point1 | 5813090752 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 21 | M_lvPNm48_R - 1975187554 | VFB_jrchk0xt | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00049779 | neuprint_JRC_Hemibrain_1point1 | 1975187554 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1451 | Z_vPNml1(noAL+SEZ)_R - 729608317 | VFB_jrchk7zr | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult antennal lobe projection neuron | FBbt_00067123 | neuprint_JRC_Hemibrain_1point1 | 729608317 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 1456 | M_vPNml63_R - 1857143769 | VFB_jrchk0yl | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00007414 | neuprint_JRC_Hemibrain_1point1 | 1857143769 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 1461 | M_vPNml66_R - 5813071409 | VFB_jrchk0yr | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00007414 | neuprint_JRC_Hemibrain_1point1 | 5813071409 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 1465 | M_lPNm11A_R - 5813044177 | VFB_jrchk0w9 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00049778 | neuprint_JRC_Hemibrain_1point1 | 5813044177 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 1468 | M_l2PNm14_R - 1723506321 | VFB_jrchk0w4 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult multiglomerular antennal lobe projection... | FBbt_00049910 | neuprint_JRC_Hemibrain_1point1 | 1723506321 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... |
339 rows × 11 columns
Fetching synaptic partners
Looks good! Next: What’s downstream of those PNs?
ds = neu.fetch_simple_connections(upstream_criteria=list(map(int, ALPNs['accession'])))
#using hemibrain regex
#ds = neu.fetch_simple_connections(upstream_criteria=neu.NeuronCriteria(bodyId=pns.bodyId.values))
ds
| bodyId_pre | bodyId_post | weight | type_pre | type_post | instance_pre | instance_post | conn_roiInfo | |
|---|---|---|---|---|---|---|---|---|
| 0 | 635062078 | 1671292719 | 390 | DP1m_adPN | lLN2T_c | DP1m_adPN_R | lLN2T_c(Tortuous)_R | {'AL(R)': {'pre': 390, 'post': 390}, 'AL-DP1m(... |
| 1 | 635062078 | 1704347707 | 326 | DP1m_adPN | lLN2T_c | DP1m_adPN_R | lLN2T_c(Tortuous)_R | {'AL(R)': {'pre': 324, 'post': 324}, 'AL-DP1m(... |
| 2 | 542634818 | 1704347707 | 322 | DM1_lPN | lLN2T_c | DM1_lPN_R | lLN2T_c(Tortuous)_R | {'AL(R)': {'pre': 322, 'post': 322}, 'AL-DM1(R... |
| 3 | 635062078 | 1640922516 | 320 | DP1m_adPN | lLN2T_e | DP1m_adPN_R | lLN2T_e(Tortuous)_R | {'AL(R)': {'pre': 317, 'post': 316}, 'AL-DP1m(... |
| 4 | 724816115 | 1670916819 | 318 | DP1l_adPN | lLN2P_a | DP1l_adPN_R | lLN2P_a(Patchy)_R | {'AL(R)': {'pre': 318, 'post': 318}, 'AL-DP1l(... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 101862 | 5901222910 | 5813086037 | 1 | DM2_lPN | None | DM2_lPN_R | None | {'LH(R)': {'pre': 1, 'post': 1}} |
| 101863 | 5901222910 | 5813095915 | 1 | DM2_lPN | KCg-m | DM2_lPN_R | KCg-m_R | {'MB(R)': {'pre': 1, 'post': 1}, 'CA(R)': {'pr... |
| 101864 | 5901222910 | 5813129316 | 1 | DM2_lPN | LHAV6a1_b | DM2_lPN_R | LHAV6a1_b_R | {'LH(R)': {'pre': 1, 'post': 1}} |
| 101865 | 5901222910 | 5901193783 | 1 | DM2_lPN | LHAV4g4_a | DM2_lPN_R | LHAV4g4_a_R | {'LH(R)': {'pre': 1, 'post': 1}} |
| 101866 | 5901222910 | 5901203780 | 1 | DM2_lPN | LHAV4g11 | DM2_lPN_R | LHAV4g11_R | {'LH(R)': {'pre': 1, 'post': 1}} |
101867 rows × 8 columns
Each row is now a connections from a single up- to a single downstream neuron. The “weight” is the number of synapses between the pre- and the postsynaptic neuron. Let’s simplify by grouping by type:
by_type = ds.groupby(['type_pre', 'type_post'], as_index=False).weight.sum()
by_type.sort_values('weight', ascending=False, inplace=True)
by_type.reset_index(drop=True, inplace=True)
by_type
| type_pre | type_post | weight | |
|---|---|---|---|
| 0 | DC3_adPN | KCg-m | 3670 |
| 1 | VM5d_adPN | KCg-m | 3219 |
| 2 | DC1_adPN | KCg-m | 3215 |
| 3 | VL2a_adPN | KCg-m | 3096 |
| 4 | DA1_lPN | KCg-m | 3078 |
| ... | ... | ... | ... |
| 40631 | M_vPNml50 | WEDPN4 | 1 |
| 40632 | M_vPNml50 | WEDPN12 | 1 |
| 40633 | M_vPNml50 | V_ilPN | 1 |
| 40634 | M_vPNml50 | VP4+VL1_l2PN | 1 |
| 40635 | Z_vPNml1 | mALD2 | 1 |
40636 rows × 3 columns
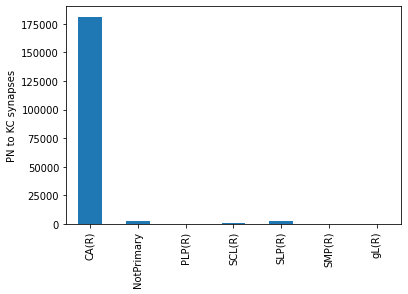
The strongest connections are between PNs and Kenyon Cells (KCs). That’s not much of a surprise since there are thousands of KCs. For the sake of the argument let’s say we want to know where these connections occur:
#First find KCs in VFB
KCs = pd.DataFrame.from_records(vc.get_instances("'adult Kenyon cell'", summary=True))
#Select only rows from Hemibrain1.1 dataset
KCs=KCs[KCs['data_source'].str.match('neuprint_JRC_Hemibrain_1point1')]
KCs
Running query: FBbt:00049825
Query URL: http://owl.virtualflybrain.org/kbs/vfb/instances?object=FBbt%3A00049825&prefixes=%7B%22FBbt%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FFBbt_%22%2C+%22RO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FRO_%22%2C+%22BFO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FBFO_%22%7D&direct=False
Query results: 5107
| label | symbol | id | tags | parents_label | parents_id | data_source | accession | templates | dataset | license | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | KCg-m_R - 692810011 | VFB_jrchjw74 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 692810011 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 2 | KCg-m_R - 477909679 | VFB_jrchjw36 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 477909679 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 5 | KCab-m_R - 5812980222 | VFB_jrchjvqk | Entity|has_image|Adult|Anatomy|has_neuron_conn... | alpha/beta Kenyon cell | FBbt_00100248 | neuprint_JRC_Hemibrain_1point1 | 5812980222 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 6 | KCg-m_R - 5812982832 | VFB_jrchjw83 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 5812982832 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 7 | KCab-m_R - 5812980806 | VFB_jrchjvr2 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | alpha/beta Kenyon cell | FBbt_00100248 | neuprint_JRC_Hemibrain_1point1 | 5812980806 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5093 | KCg-m_R - 664184060 | VFB_jrchjwck | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 664184060 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 5096 | KCg-m_R - 5812982620 | VFB_jrchjwdd | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 5812982620 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 5100 | KCg-m_R - 661430179 | VFB_jrchjwdb | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 661430179 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 5101 | KCg-m_R - 5812982436 | VFB_jrchjwcz | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 5812982436 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 5102 | KCg-m_R - 663834152 | VFB_jrchjwa9 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | gamma main Kenyon cell | FBbt_00111061 | neuprint_JRC_Hemibrain_1point1 | 663834152 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... |
1927 rows × 11 columns
adj, roi_info2 = neu.fetch_adjacencies(sources=list(map(int, ALPNs['accession'])), targets=list(map(int, KCs['accession'])))
roi_info2.head()
0%| | 0/2 [00:00<?, ?it/s]
| bodyId_pre | bodyId_post | roi | weight | |
|---|---|---|---|---|
| 0 | 542634818 | 301314208 | CA(R) | 6 |
| 1 | 542634818 | 331999156 | CA(R) | 1 |
| 2 | 542634818 | 332344592 | CA(R) | 2 |
| 3 | 542634818 | 332344908 | CA(R) | 9 |
| 4 | 542634818 | 332353106 | CA(R) | 13 |
# Group by region of interest (ROI)
by_roi = roi_info2.groupby('roi').weight.sum()
by_roi.head()
roi
CA(R) 181274
NotPrimary 2764
PLP(R) 11
SCL(R) 509
SLP(R) 2026
Name: weight, dtype: int64
ax = by_roi.plot.bar()
ax.set_xlabel('')
ax.set_ylabel('PN to KC synapses')
Text(0, 0.5, 'PN to KC synapses')
Querying paths
Let’s say we want to find out how to go from a PN (second order olfactory neurons) all the way to a descending neuron (presumably leading to motor neurons in the VNC).
#First find DNs in VFB
DNs = pd.DataFrame.from_records(vc.get_instances("'adult descending neuron'", summary=True))
#Select only rows from Hemibrain1.1 dataset
DNs=DNs[DNs['data_source'].str.match('neuprint_JRC_Hemibrain_1point1')]
DNs
Running query: FBbt:00047511
Query URL: http://owl.virtualflybrain.org/kbs/vfb/instances?object=FBbt%3A00047511&prefixes=%7B%22FBbt%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FFBbt_%22%2C+%22RO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FRO_%22%2C+%22BFO%22%3A+%22http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FBFO_%22%7D&direct=False
Query results: 139
| label | symbol | id | tags | parents_label | parents_id | data_source | accession | templates | dataset | license | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | DNp27_R - 1072063538 | VFB_jrchjthm | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the posterior brain DNp27 | FBbt_00047660 | neuprint_JRC_Hemibrain_1point1 | 1072063538 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 3 | DNp16/17_R - 1655626125 | VFB_jrchjth0 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the posterior brain | FBbt_00047517 | neuprint_JRC_Hemibrain_1point1 | 1655626125 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 7 | DNb04_R - 1037393225 | VFB_jrchjtfx | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the anterior ventral brai... | FBbt_00047585 | neuprint_JRC_Hemibrain_1point1 | 1037393225 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 9 | DNp32_R - 5813050455 | VFB_jrchjthr | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the posterior brain DNp32 | FBbt_00047665 | neuprint_JRC_Hemibrain_1point1 | 5813050455 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 11 | NPFP1_R - 1043117106 | VFB_jrchk103 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult NPF P1 neuron | FBbt_00048120 | neuprint_JRC_Hemibrain_1point1 | 1043117106 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 133 | DNp17_R - 1498383456 | VFB_jrchjth4 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the posterior brain DNp17 | FBbt_00047650 | neuprint_JRC_Hemibrain_1point1 | 1498383456 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 134 | DNg30_R - 571346836 | VFB_jrchjtg4 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the gnathal ganglion DNg30 | FBbt_00047622 | neuprint_JRC_Hemibrain_1point1 | 571346836 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 135 | DNp27_L - 5813020347 | VFB_jrchjthn | Entity|has_image|Adult|Anatomy|has_neuron_conn... | descending neuron of the posterior brain DNp27 | FBbt_00047660 | neuprint_JRC_Hemibrain_1point1 | 5813020347 | JRC_FlyEM_Hemibrain|JRC2018Unisex | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 137 | MDN(PDM13)_L - 5813021079 | VFB_jrchk0vf | Entity|has_image|Adult|Anatomy|has_neuron_conn... | adult moonwalker descending neuron | FBbt_00111308 | neuprint_JRC_Hemibrain_1point1 | 5813021079 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... | |
| 138 | Giant Fiber_R - 2307027729 | VFB_jrchjup1 | Entity|has_image|Adult|Anatomy|has_neuron_conn... | giant fiber neuron | FBbt_00004020 | neuprint_JRC_Hemibrain_1point1 | 2307027729 | JRC2018Unisex|JRC_FlyEM_Hemibrain | Xu2020NeuronsV1point1 | https://creativecommons.org/licenses/by/4.0/le... |
101 rows × 11 columns
# First fetch the DNs
dns, _ = neu.fetch_neurons(list(map(int, DNs['accession'])))
#with regex
#dns, _ = neu.fetch_neurons(neu.NeuronCriteria(type='(.*DN[^1]{0,}.*|Giant Fiber)', regex=True))
dns
| bodyId | instance | type | pre | post | size | status | cropped | statusLabel | cellBodyFiber | somaRadius | somaLocation | inputRois | outputRois | roiInfo | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 451689001 | DNp25_R | DNp25 | 165 | 803 | 948309595 | Traced | False | Roughly traced | PDM05 | 361.0 | [22185, 13374, 16352] | [ATL(R), CA(R), FLA(R), INP, LH(R), MB(+ACA)(R... | [ATL(R), CA(R), INP, MB(+ACA)(R), MB(R), SLP(R... | {'SNP(R)': {'pre': 158, 'post': 703, 'downstre... |
| 1 | 512851433 | DNp10(PDM27)_L | DNp10 | 25 | 2487 | 3010901036 | Traced | False | Roughly traced | None | 483.5 | [27334, 21053, 7640] | [ATL(L), GC, GOR(L), IB, ICL(L), INP, SCL(L), ... | [ATL(L), GC, GOR(L), ICL(L), INP, SCL(L), SMP(... | {'SNP(L)': {'pre': 7, 'post': 512, 'downstream... |
| 2 | 519949044 | oviDNb_R | oviDNb | 7 | 1089 | 1618057638 | Traced | False | Roughly traced | PDM15 | 396.0 | [19291, 15307, 11696] | [AVLP(R), CA(R), CRE(-ROB,-RUB)(R), CRE(-RUB)(... | [CRE(-RUB)(L), CRE(L), INP, SIP(R), SMP(L), SM... | {'SNP(R)': {'pre': 2, 'post': 656, 'downstream... |
| 3 | 542751938 | DNp44_R | DNp44 | 182 | 634 | 1674610618 | Traced | False | Roughly traced | PVM09 | 321.0 | [18875, 11698, 13136] | [AL(R), AL-VL1(R), AL-VL2p(R), AL-VP1d(R), AL-... | [AL(R), AL-DP1m(R), AL-VL1(R), AL-VL2p(R), AL-... | {'SNP(R)': {'pre': 21, 'post': 277, 'downstrea... |
| 4 | 550655668 | oviDNa_R | oviDNa | 36 | 1111 | 1887226051 | Traced | False | Roughly traced | PDM15 | 301.0 | [19199, 15756, 13456] | [AOTU(R), CRE(-ROB,-RUB)(R), CRE(R), EPA(R), I... | [CRE(-ROB,-RUB)(R), CRE(R), INP, SCL(R), SIP(R... | {'SNP(R)': {'pre': 14, 'post': 772, 'downstrea... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | 5813068915 | NPFP1(PDM12)_L | NPFP1 | 123 | 108 | 4221447995 | Traced | False | Roughly traced | None | 539.5 | [28242, 20366, 6808] | [ATL(L), AVLP(R), CAN(R), EPA(R), GOR(R), IB, ... | [AVLP(R), CAN(R), EPA(R), GOR(R), IB, ICL(L), ... | {'INP': {'pre': 34, 'post': 47, 'downstream': ... |
| 97 | 5813077405 | DNd01_L | DNd01 | 21 | 731 | 738596888 | Traced | False | Roughly traced | None | 342.0 | [25173, 33207, 14176] | [SMP(L), SMP(R), SNP(L), SNP(R)] | [SMP(L), SMP(R), SNP(L), SNP(R)] | {'SNP(L)': {'pre': 13, 'post': 675, 'downstrea... |
| 98 | 5813078134 | DNp15_R | DNp15 | 21 | 2310 | 1288564222 | Traced | False | Roughly traced | None | NaN | None | [CAN(R), GNG, IPS(R), PENP, SPS(R), VLNP(R), V... | [GNG, IPS(R), VMNP] | {'VMNP': {'pre': 7, 'post': 860, 'downstream':... |
| 99 | 5813095193 | DNp16_R | DNp16 | 70 | 683 | 292910198 | Traced | False | Roughly traced | PVM02 | 276.0 | [25941, 10730, 20000] | [CAN(R), GNG, IB, INP, IPS(R), PENP, SPS(R), V... | [CAN(R), GNG, IPS(R), PENP, SPS(R), VMNP] | {'INP': {'post': 34, 'upstream': 34}, 'IB': {'... |
| 100 | 5813108230 | DNp16/17_R | DNp16/17 | 5 | 177 | 304268206 | Traced | False | Roughly traced | PVM02 | 321.5 | [23677, 10111, 18544] | [GNG, IB, INP, IPS(R), SPS(R), VMNP] | [IPS(R), SPS(R), VMNP] | {'INP': {'post': 2, 'upstream': 2}, 'IB': {'po... |
101 rows × 15 columns
Neuprint lets you query paths from a single source to a single target. For multi-source or -target queries, your best bet is to download the entire graph and run the queries locally using networkx or igraph.
# Find all paths from A PN to A DNs
paths = neu.fetch_shortest_paths(upstream_bodyId=list(map(int, ALPNs['accession']))[0],
downstream_bodyId=list(map(int, DNs['accession']))[0],
min_weight=10)
paths
| path | bodyId | type | weight | |
|---|---|---|---|---|
| 0 | 0 | 294792184 | M_vPNml53 | 0 |
| 1 | 0 | 5813057148 | SLP387 | 16 |
| 2 | 0 | 511349908 | SMP183 | 19 |
| 3 | 0 | 545501062 | SMP464 | 10 |
| 4 | 0 | 739256609 | CL029 | 25 |
| ... | ... | ... | ... | ... |
| 343 | 57 | 5813057148 | SLP387 | 16 |
| 344 | 57 | 357515217 | SLP365 | 12 |
| 345 | 57 | 479607277 | CL102 | 10 |
| 346 | 57 | 449250041 | CL026 | 22 |
| 347 | 57 | 1072063538 | DNp27 | 22 |
348 rows × 4 columns
So it looks like there are three separate 7-hop paths to go from M_vPNml53 to DN1a. Let’s visualize the neurons involved!
Fetching meshes & skeletons
You can fetch skeletons as SWCs directly via neuprint-python. For visualization however it’s easiest to load neuron morphologies via navis. For that navis wraps neuprint-python and adds some convenience functions (see also the tutorial):
# Import the wrapped neuprint-python
# -> this exposes ALL base functions plus a couple navis-specific extras
import navis
import navis.interfaces.neuprint as neu
#client = neu.Client('https://neuprint.janelia.org', dataset='hemibrain:v1.1')
# Fetch neurons in the first path
nl = neu.fetch_skeletons(paths.loc[(paths.path == 0), 'bodyId'])
nl
| type | name | id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | navis.TreeNeuron | M_vPNml53_R | 294792184 | 3670 | 436 | 180 | 190 | 1.877807e+05 | 3649 | 8 nanometer |
| 1 | navis.TreeNeuron | SMP464_R | 545501062 | 4507 | 454 | 183 | 193 | 1.888869e+05 | 4 | 8 nanometer |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4 | navis.TreeNeuron | DNp27_R | 1072063538 | 47115 | 9298 | 1750 | 1888 | 2.720497e+06 | 25808 | 8 nanometer |
| 5 | navis.TreeNeuron | CL029_R | 739256609 | 31953 | 7932 | 2660 | 2709 | 1.275071e+06 | 6033 | 8 nanometer |
# Let's also get some ROI meshes
al = neu.fetch_roi('AL(R)')
lh = neu.fetch_roi('LH(R)')
ca = neu.fetch_roi('CA(R)')
# Plot
navis.plot3d([nl, lh, al, ca], width=1100)
Plotting graphs
There are various ways of plotting static graphs. In theory Jupyter notebooks lend themselves to interactive graphs too but unfortunately DeepNote does not yet support the required libraries (e.g. ipywidgets). That being said: if you want to run this locally or on Google colab, check out ipycytoscape.
There are numerous options to do this but we will use networkx to plot a static graph:
import networkx as nx
import numpy as np
# Initialize the graph
G = nx.DiGraph()
# Generate edges from the paths
edges = []
for p in paths.path.unique():
this_path = paths.loc[(paths.path == p)]
this_edges = list(zip(this_path.values[:-1], this_path.values[1:]))
for i in range(this_path.shape[0] - 1):
edges.append([this_path.bodyId.values[i], this_path.bodyId.values[i + 1], this_path.weight.values[i + 1]])
# Add the edges
G.add_weighted_edges_from(edges)
# Add some names to the nodes
nx.set_node_attributes(G, paths.set_index('bodyId')['type'].to_dict(), name='name')
import matplotlib.pyplot as plt
# Draw using a simple force-directed layout
pos = nx.kamada_kawai_layout(G)
# We could draw everything in one step but this way we have more control over the plot
fig, ax = plt.subplots(figsize=(10, 10))
# Draw nodes
nx.draw_networkx_nodes(G, pos=pos, ax=ax)
# Draw edges
weights = np.array([e[2]['weight'] for e in G.edges(data=True)])
nx.draw_networkx_edges(G, pos=pos, width=(weights / 12).tolist())
# Add node labels
nx.draw_networkx_labels(G, pos=pos, labels=dict(G.nodes('name')), font_size=14)
# Turn axes of
ax.set_axis_off()

In general, I recommend exporting your graph to e.g. graphml and importing it into e.g. cytoscape if you want to explore an interactive network graph.
nx.write_gml(G, "my_graph.gml")
Feedback
Was this page helpful?
Glad to hear it! Please tell us how we can improve.
Sorry to hear that. Please tell us how we can improve.